SUMMARY
Some Ts in nuclear DNA of trypanosomes and Leishmania are hydroxylated and glucosylated to yield base J (β-D-glucosyl-hydroxymethyluracil). In Leishmania, about 99% of J is located in telomeric repeats. We show here that most of the remaining J is located at chromosome-internal RNA polymerase II termination sites. This internal J and telomeric J can be reduced by a knockout of J-binding protein 2 (JBP2), an enzyme involved in the first step of J biosynthesis. J levels are further reduced by growing Leishmania JBP2 knockout cells in BrdU-containing medium, resulting in cell death. The loss of internal J in JBP2 knockout cells is accompanied by massive readthrough at RNA polymerase II termination sites. The readthrough varies between transcription units but may extend over 100 kb. We conclude that J is required for proper transcription termination and infer that the absence of internal J kills Leishmania by massive readthrough of transcriptional stops.
INTRODUCTION
In 1993, a new DNA base, β-D-glucosyl-hydroxymethyluracil (base J), was discovered in the nuclear DNA of the African trypanosome Trypanosoma brucei (Gommers-Ampt et al., 1993). Base J replaces about 1% of thymine (T) in the genome and is predominantly present in repetitive DNA, such as telomeric repeats (Gommers-Ampt et al., 1993; van Leeuwen et al., 1996, 1997, 2000). Base J was subsequently found in the DNA of all other kinetoplastid flagellates tested (van Leeuwen et al., 1998b) and in the related unicellular alga Euglena gracilis (Dooijes et al., 2000), but not in other organisms, making biosynthesis of J a potential target for chemotherapy of pathogenic kinetoplastids (Borst and Sabatini, 2008).
Indirect evidence, reviewed in Borst and Sabatini (2008), indicates that J is synthesized in two steps: in the first step, T residues at specific positions in DNA are hydroxylated to form hydroxymethyluracil (HOMeU); in the second step, a glucose is attached to HOMeU, yielding J. Two enzymes are thought to catalyze the first step. The first was initially identified as a protein specifically binding to duplex DNA containing J (Cross et al., 1999). This J-binding protein 1 (JBP1) was later found to affect J levels (Cross et al., 2002) and to contain a thymidine hydroxylase domain (Yu et al., 2007). The second putative hydroxylase, J-binding protein 2 (JBP2), does not bind to J-DNA (DiPaolo et al., 2005); in addition to its thymidine hydroxylase domain (DiPaolo et al., 2005; Yu et al., 2007; Vainio et al., 2009), it contains a SWI/SNF domain essential for its activity (DiPaolo et al., 2005). Both JBP1 and JBP2 appear to belong to the new TET/JBP subfamily of dioxygenases requiring Fe2+ and 2-oxoglutarate for activity (Iyer et al., 2009; Tahiliani et al., 2009).
Whereas T. brucei can survive without J (Cliffe et al., 2009, 2010), Leishmania tarentolae, a pathogen of lizards easily cultured in the lab, appears to require J because JBP1 knockout (KO) cells are not viable (Genest et al., 2005). The KO of JBP2 initially did not result in detectable defects, but these cells lose J; after >30 passages, only about 30% of J remains (Vainio et al., 2009). When these cells are grown in bromodeoxyuridine (BrdU), they lose more J and die (Vainio et al., 2009). The cause of this BrdU-induced J loss, previously found in T. brucei (van Leeuwen et al., 1998a), is not known, but it provides a tool to test possible functions of base J. In Leishmania species, >99% of all J is present in telomeric repeats (Genest et al., 2007), and we therefore looked for a telomeric phenotype in L. tarentolae JBP2 KO cells dying in BrdU. None were found. No other clue to the cause of death was found either. The dying cells do not stop in a defined phase of the cell cycle; they do not activate a DNA damage response (P.B., S.V., P.-A.G., and H.G.A.M.v.L., unpublished data).
This deadlock was broken by Cliffe et al. (2010), who discovered that a minor fraction of J in T. brucei has a well-defined chromosome-internal distribution. High-throughput sequencing confirmed the location of J in repetitive DNA but, in addition, revealed chromosome-internal J peaks coinciding with RNA polymerase (RNAP) II transcriptional initiation and termination sites (Cliffe et al., 2010). The transcriptome of Leishmania and other kinetoplastids is unusual with RNAP-II-directed polycistronic transcription of long clusters of protein-coding genes (El-Sayed et al., 2005b; Berriman et al., 2005; Jackson et al., 2010). Mature messenger RNAs (mRNAs) are produced from the primary transcripts by trans-splicing and polyadenylation. Trans-splicing adds a capped 35 nucleotide sequence to the 5′ end of each mRNA (Boothroyd and Cross, 1982; Van der Ploeg et al., 1982; De Lange et al., 1983; Nelson et al., 1983; Sutton and Boothroyd, 1986; Agabian, 1990). This spliced leader (SL) or miniexon sequence is derived from a precursor transcribed by RNAP II from a separate set of SL precursor genes (Kooter and Borst, 1984; De Lange et al., 1984). The ribosomal RNA (rRNA) (transcribed by RNAP I), tRNA, 5S RNA, and snRNA genes (transcribed by RNAP III) are scattered between RNAP II polycistronic transcription units (PTUs) (Padilla-Mejía et al., 2009).
Although 99% of all J in L. tarentolae is in telomeric repeats (Genest et al., 2007), we have shown that the remaining J is in chromosome-internal positions, but we were unable to map this internal J by standard cloning procedures (Genest et al., 2007). We have now confirmed the presence of internal J at RNAP II transcription termination sites in L. major and L. tarentolae. We show that this internal J is strongly reduced in JBP2 KO L. tarentolae cells. This loss of J is accompanied by massive readthrough of normal RNAP II transcriptional termination sites.
RESULTS
Localization of Internal J in the Genome of L. major
For the initial mapping of internal J in Leishmania, we used L. major because the genomic map of this species is the most refined (Myler et al., 1999; Ivens et al., 2005) and has been used for mapping transcripts (Myler et al., 2000; Belli et al., 2003; Worthey et al., 2003; Martínez-Calvillo et al., 2003, 2004; Monnerat et al., 2004) and chromatin modifications (Thomas et al., 2009). The DNA of the L. major Friedlin strain was sheared, and J-containing fragments were enriched by immunoprecipitation with a specific anti-J-DNA antiserum (van Leeuwen et al., 1997) or by binding of the DNA fragments to Strep(II)-tagged JBP1 and retrieval of the bound fragments with streptavidin beads (see Experimental Procedures). Both methods enriched J-containing DNA fragments effectively, as shown by the high signal-to-noise ratio of the results presented here (see Figure 1 and Data S1 available online). To simplify the presentation, we mainly show the data obtained with the anti-J-DNA antiserum, but all J peaks discussed here were also found by using the Strep(II)-tagged JBP1.
Figure 1. Location of Base J in the L. major and L. tarentolae Genomes.
(A–D) The location of base J in the L. major (A–C) and L. tarentolae (D) genomes was mapped by deep sequencing. DNA fragments enriched for J were isolated by either an immunoprecipitation with anti-J-DNA antibodies or by a pull-down with JBP1, sequenced, and mapped. As an example, we show the location of J on chromosome 14 of L. major (A) and L. tarentolae (D). All L. major and L. tarentolae chromosomes are presented in Data S1. The upper row in (A) shows the genomic map of L. major chromosome 14 with the blue bars representing the protein coding regions organized in long PTUs encoded by the top (transcription to the right) or bottom strand (transcription to the left). The arrows indicate the transcriptional orientation of the PTUs. The green bars represent RNA genes. The next row shows the DNA segments immunoprecipitated with an antiserum against acetylated histone H3 (Thomas et al., 2009), marking putative transcription start sites. Underneath are two rows showing the localization and abundance (as the total number of reads within 100 bp consecutive bins) of J-containing fragments over the entire chromosome by deep sequencing using either anti-J-DNA antiserum or Strep(II)-tagged JBP1 to enrich for J-containing DNA fragments. Red bars indicate values going off-scale. (B) shows an example of an internal J peak between two histone H3ac peaks in a divergent transcription start region on chromosome 7 of L. major, and (C) shows an internal J peak upstream of a histone H3ac peak within a PTU on chromosome 4. (D) shows the genomic organization of L. tarentolae chromosome 14 and the localization and abundance of base J (using anti-J-DNA antiserum). See also Data S1 and Tables S1 and S2.
Our high-throughput sequencing locates the bulk of J in telomeric repeats, confirming earlier analyses (Genest et al., 2007). In addition, we find internal J at three main locations. Prominent internal J peaks occur where two RNAP II PTUs converge and terminate (see Figure 1A for an example on chromosome 14). Of the 38 convergent transcription termination regions in the L. major genome, all but one had a J peak. A second location where internal J peaks are found is where RNAP II transcription starts in both directions. Although not all of these start regions contain internal J (see Figure 1A and Data S1), 16 of the 58 do contain a major internal J peak (see Figure 1B for an example on chromosome 7). These divergent transcription start regions are marked by two adjacent acetylated histone H3 (H3ac) peaks (Thomas et al., 2009), and the internal J peak is invariably found between these H3ac peaks. The size of the divergent start region (as measured by the distance between paired H3ac peaks) correlates with the presence of J because the 12 largest all have a major J peak. Conversely, most start regions with H3ac peaks close together have no J peak. A third major location of J peaks is immediately adjacent to single H3ac peaks within PTUs (see Figure 1C for an example on chromosome 4). The relative location of the internal J and H3ac peaks is what one would expect if these J peaks mark transcription termination sites and the H3ac peaks represent a restart of transcription in the same direction. In addition, scattered internal J peaks are found throughout the genome. We assume that their appearance in the middle of PTUs is an unavoidable mapping artifact due to inaccuracies in the current genomic map. Modest J peaks were also observed upstream of the SL RNA locus in chromosome 2 and flanking the rRNA gene clusters in chromosome 27.
Loss of Internal J Is Concomitant with Overall J Loss in L. tarentolae
To study the effect of J loss, we switched to L. tarentolae, as all mutants affecting J levels were made in L. tarentolae. Although the assembly of the L. tarentolae genome is not yet as refined as that of L. major, the overall genome organization of the two species is very similar and so is the location of J. In fact, all major internal J peaks in L. major can be found at corresponding positions in the L. tarentolae genome (see Figures 1A and 1D and Data S1). As we have far more reads on J-DNA fragments from L. tarentolae than from L. major, we also see more minor J peaks. For instance, for the 58 divergent transcription start regions of L. tarentolae, we find a major J peak in 22, a minor J peak in 18, and no J in 26.
We have previously shown that disruption of JBP2, one of the genes involved in J biosynthesis, results in a gradual loss of J (Vainio et al., 2009). When these JBP2 KO cells are cultured in BrdU-containing medium (KO+BrdU), they lose additional J and die (Vainio et al., 2009). We now find that the overall drop in J levels in L. tarentolae JBP2 KO is accompanied by a profound decrease in internal J, as determined by high-throughput sequencing of J-containing fragments (see Figure 2 and Data S2). Under the conditions of these experiments, the overall J levels in JBP2 KO cells, measured by quantitative dot blots, varied between 30% and 37% of wild-type (WT) (n = 3). For the corresponding samples of JBP2 KO L. tarentolae grown for 4 or 5 days in KO+BrdU, the J levels were 13%–16% of WT. Nearly all of this J is present in telomeric repeats (Genest et al., 2007), and indeed, these repeats are still prominently precipitated in the KO+BrdU DNA samples (Data S2). In contrast, the decrease in internal J looks far more pronounced than that of overall J because many internal J peaks are substantially reduced in the KO cells and disappear almost completely in the KO+BrdU cells, implying >99% loss of internal J. However, as the dot blots and the IPs are both semiquantitative (van Leeuwen et al., 1997), the decrease illustrated in Figure 2 (and Data S2) cannot be interpreted in a strictly quantitative fashion.
Figure 2. Loss of J in L. tarentolae Leads to Transcriptional Readthrough.
The location of protein coding units, RNA genes, the direction of transcription of the PTUs, and the location and abundance of base J on chromosome 30 of L. tarentolae are presented as in Figure 1. Bars going up indicate the number of transcripts derived from the top strand; bars going down indicate the number of transcripts derived from the bottom strand. J levels and small RNAs are plotted on a linear scale in bins of 100 bp, and the levels of processed RNAs are plotted on a logarithmic scale also in bins of 100 bp. Red bars are off-scale. The broken vertical line marks the transcription start and stop sites. The RNA samples were normalized by the total number of reads in each library. Data S2 presents the results for all L. tarentolae chromosomes. See also Data S2 and Tables S1 and S2.
Loss of J Results in Massive Readthrough of RNAP II Transcription Units
Because internal J is predominantly found at transcription termination sites and is lost from these sites in JBP2 KO mutants, we tested whether the loss of J affects transcription termination. As synthesis of mature mRNAs requires processing sequences that might be underrepresented in abnormal RNA produced by readthrough of termination sites, we initially looked at the small RNA fraction, as this might contain a complete set of antisense RNA degradation products. The high-throughput RNA sequencing method used in this analysis will capture all RNA degradation products with 5′-P and 3′-OH ends. Because Leishmania appears to have few (if any) microRNAs (miRNAs), most sequences obtained by this analysis will be derived from degradation of (pre-)mRNAs.
More than 99% of the small RNAs found in WT cells correspond to the coding strand predicted from the preliminary map of L. tarentolae (see Table S1). The result is a rough picture of the primary transcript map of L. tarentolae, as reflected in small degradation products (see Figure 2 and Data S2). However, there is a striking change in strand specificity in the JBP2 KO mutant (where 7% of all the small RNAs are antisense), and this is exacerbated when these cells are grown in the presence of BrdU (KO+BrdU) (9.6% is antisense). We attribute this profound change primarily to readthrough of convergent transcription termination regions, when these regions lose their J, rather than false starts on the wrong strand. First, the area between protein-coding genes at large convergent transcription termination regions fills up with transcripts from both strands (see Data S2 for an example on chromosome 14). Second, the frequency of antisense small RNA fragments decreases with increasing distance from the convergent transcription termination region. This is illustrated in Figure 3A, in which we plot the average transcript level on the same strand before and after the transcription stop site. Whereas this level drops to near zero in RNA from WT cells directly after the transcription stop site, it remains high in the JBP2 KO and KO+BrdU samples and then gradually tapers off. Finally, the amount and extent of readthrough is generally inversely related to the amount of J at the convergent transcription termination regions; i.e., the more J is reduced, the more readthrough is seen and the further it extends from the transcription stop site (see Data S2).
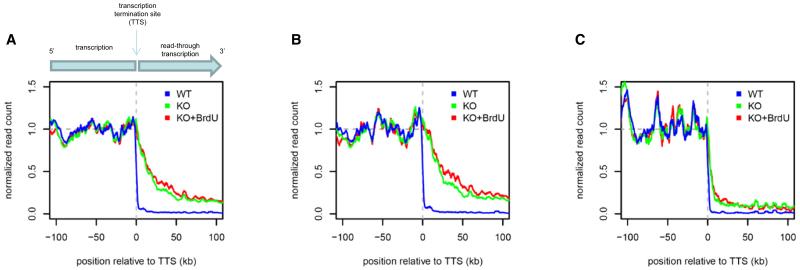
Figure 3. Transcription Readthrough upon Loss of JBP2.
(A) Average PTU readthrough in JBP2−/− (KO, green) and JBP2−/− grown in the presence of BrdU (KO+BrdU, red) as compared to WT (blue). Curves show running means of small RNA read counts across the transcription termination sites for all convergent PTUs, aligned by their stop sites, taking transcript orientation into account. Negative x axis coordinates represent positions upstream of the stop site within the PTUs, and positive coordinates are downstream of the stop site. The median read density within each PTU was adjusted to unity; hence, downstream transcription levels are values relative to intra-PTU levels.
(B and C) The presence of oppositely oriented RNAP III genes in the convergent transcription termination regions drastically decreases readthrough. Similar plots as in (A), but now only for PTUs without RNAP III genes in corresponding convergent transcription termination regions or with RNAP III genes in the corresponding convergent transcription termination regions on the same strand as the upstream PTU (B) and for PTUs with RNAP III genes in the corresponding convergent transcription termination regions on the opposite strand (C). See also Figure S1.
The transcript analysis of JBP2 KO cells presented here was done with a KO clone analyzed >100 passages after its generation. This clone had lost nearly all internal J. To test whether a more limited loss of internal J would also lead to (more modest) readthrough, we made use of our observation that the JBP2 KO cells lose J over time (Vainio et al., 2009). We isolated a fresh JBP2−/− clone and analyzed overall J, internal J, and readthrough at the earliest time points (Figure S1). Although the P0 cells had already lost 20%–30% of overall J, the loss of internal J at convergent transcription termination regions was on average only 20% and varied between undetectable and 50%. Nevertheless, very modest readthrough was already detectable at some of these early time points (Figure S1). These results suggest that physiological levels of internal J are essential for proper control of transcription termination and that even a small decrease results in readthrough.
Stable Antisense Transcripts
To test how loss of J affects the levels of mature mRNAs, we analyzed all RNAs containing the 5′-SL sequence by deep sequencing (SL or processed RNA). In WT cells, the vast majority (>99%) of SL-containing reads were restricted to the coding strand (see Table S1). The antisense reads increased considerably in the JBP2 KO mutant and further increased when this mutant was grown in the presence of BrdU (see Figure 2 and Data S2). However, the proportion of antisense SL RNA sequencing (RNA-seq) reads was lower than for small RNA-seq reads, and they were less evenly distributed, although the number of reads at each site was substantially larger than for the small RNA reads. Hence, the readthrough reflected in small RNA is often more prominent than that in SL RNA (see Figure 2 and Data S2).
The Effect of RNAP III Transcription on Readthrough
Although readthrough occurs at most convergent transcription termination regions when J levels fall, the extent of readthrough varies, and it is often asymmetric at individual termination sites (see Data S2). The presence of RNA genes that are transcribed by RNAP III (such as transfer RNAs [tRNAs] and small nuclear RNAs [snRNAs]) in the strand-switch region is the main factor determining these variations (Figures 3 and 4). When there are no RNAP III genes at the transcription termination site or RNA genes read in the same direction as the adjacent Pol II PTU, readthrough is extensive. Conversely, if RNAP II meets RNAP III head on, readthrough is modest. These results suggest that RNAP II can pass an RNAP III transcription unit more easily if the polymerases are traveling in the same direction. The readthrough at individual transcription stop sites is variable, as illustrated in Figure 4A. The profound readthrough at transcription stop sites without RNAP III genes is clear from Figure 4B, which plots the ratio of antisense over sense small RNA. The ratio of antisense over sense reads in the readthrough area often reaches or exceeds unity. This is true at some convergent transcription termination regions, even for the SL RNA-seq reads (data not shown). It is remarkable that antisense RNA is processed so efficiently into SL RNA, suggesting that polypyrimidine sequences required for SL addition and polyadenylation are relatively frequent in random RNA sequences and that these processing signals are selected against in sense RNA to allow proper mRNA formation.
Figure 4. Transcriptional Readthrough of Individual RNAP II Termination Sites at Convergent Transcription Termination Regions.
(A) Antisense reads mapping adjacent to convergent transcription termination regions. The number of antisense reads of small RNA mapping to the 20 kb window (or less for shorter PTUs) upstream (left side) or downstream (right side) of the convergent termination site was divided by the window length (in kb) and expressed per million reads for each small RNA-seq library of WT, KO, and KO+BrdU cultures. The convergent transcription termination regions are clustered in four groups with RNAP III genes in the convergent termination region on both strands (T and B), on the top stand only (T), on the bottom strand only (B), or with no RNA genes. Each row represents one transcription termination region. Arrows at the top of bars indicate values that are off-scale. The numbers in the middle refer to the detailed maps of the termination regions listed in Table S4.
(B) The ratio of antisense to sense reads from the small RNA-seq libraries adjacent to the convergent transcription termination sites. This analysis uses the same windows and symbols as in (A). See also Tables S3 and S4.
Whereas the presence of internal J and RNAP III transcription units appears to be the dominant factor in preventing the readthrough observed, there are additional variations in the extent and/or magnitude of readthrough. For example, not all convergent transcription termination regions lacking RNA genes show readthrough in both directions (see 10.2 in Figure 4), whereas some termination regions with RNA genes show readthrough in the direction opposite to that of RNAP III transcription (see 05.2 and 03.3 in Figure 4). These differences could be due to relative promoter strength, unrecognized RNAP III transcription units, or the presence of cryptic terminators not related to J.
Readthrough in WT L. tarentolae at the Only J-less Convergent Transcription Termination Region
The only convergent transcription termination region without detectable J in WT cells is present around position 580,000 on chromosome 28 (Figure 5). In this figure, we present the data as the proportion of sense to antisense small RNA-seq reads in bins of 100 bp across the chromosome to accentuate the antisense RNA. It is obvious that the J-less convergent transcription termination region shows substantial readthrough in WT cells (also visible for chromosome 28 in Data S2 and convergent transcription termination region 28.2 in Figure 4), supporting our conclusion that J at transcription termination sites is needed for proper termination. Despite the apparent absence of J at this convergent transcription termination site in WT cells, readthrough appears to increase in L. tarentolae JBP2 KO (±BrdU) mutants. Why a J-less stop should be affected by loss of J elsewhere in the genome is not clear, and we return to this point in the Discussion.
Figure 5. Location of J and Small RNAs on Chromosome 28, which Contains the Only Convergent Transcription Termination Region without J in WT L. tarentolae.
The presentation of protein coding regions and J location is as in Figure 1. The small RNA sequence data are shown by vertical bars whose ends represents the fraction of top-strand (values between 0 and 1) and bottom-strand (values between 0 and −1) reads in bins of 100 bp across the chromosome. The total number of reads in each bin is indicated by the color of the bar, with warmer (i.e. redder) colors representing more reads per bin. See Data S2 for a quantitative presentation of sense and antisense RNA.
Antisense Transcripts at Transcriptional Start Regions
Although the bulk of antisense reads appears around convergent transcription termination regions when J levels drop, there is also an increase in antisense RNA at many transcription start sites, as can be seen in Data S2. This increase is especially obvious close to the transcription start site, as illustrated in Figure 6 in which we plot the antisense reads over the entire PTU and in 5 kb windows at the beginning and end of each PTU, as explained in Figure 6A. There is also a substantial increase in antisense reads in the first 5 kb of the PTU, especially in those divergent start regions losing base J. This is especially pronounced in the KO+BrdU samples (Figure 6B), in which the level of antisense SL RNA rises to the levels found at convergent transcription termination regions. This appears to be due to an increase in erroneous starts within and around divergent transcription start regions rather than readthrough from adjacent PTUs because it is also observed at divergent start regions with no upstream convergent transcription termination regions (e.g. the left end of chromosome 30 in Figure 2). In contrast, the increase in antisense RNA at transcription start sites is more modest when measured by small RNA-seq (Figure 6C). This difference could be due to enrichment of processed RNAs by the SL RNA-seq method. Trans-splicing takes place during transcription and favors the use of the most proximal SL acceptor site (Ullu et al., 1993); this site is mapped by using the SL RNA-seq method. Every false transcription start upstream of a PTU will result in SL RNAs mapping upstream of this PTU, scored as antisense RNA. Small RNA-seq, on the other hand, maps RNA degradation products derived from the entire transcript, and these may largely overlap with sense transcripts, as indicated in Figure 6A.
Figure 6. Effects of Knocking Out the JBP2 Gene and Growth in BrdU of L. tarentolae on Antisense Transcript Levels.
(A–C) Antisense transcripts due to premature starts or readthrough were quantified in WT, KO, and KO+BrdU cells. (A) shows a schematic representation of the antisense quantification due to false starts (left side) or readthrough (right side). Normal (WT) transcription of the PTUs (blue bars) is indicated by the black arrows; when base J is lost, J transcription initiation or termination is compromised, leading to transcripts starting upstream of the PTU or continuing downstream of the PTU, as indicated by the red arrows. This is detected as antisense RNA in the PTU on the other side of the transcription start site (false starts) or transcription termination site (readthrough). The antisense SL or small RNA mapped to the first or last 5 kb of the PTU on the other side of the transcription termination site is used to measure, respectively, premature starts or readthrough (dashed red arrows). (B) shows the transcript levels of antisense RNA based on SL RNA-seq and (C) on small RNA-seq (the combined values of three independent experiments). Read densities are expressed per kb normalized to one million reads. The first block of bars in (B) and (C) represents the mean antisense read density over the entire PTU for all 183 PTUs. The effect of false starts is represented as the mean antisense RNA read density in the first 5 kb of all PTUs next to a divergent transcription start site (second block) or of all 22 PTUs next to a divergent start site with a major J peak (third block). The fourth block is the mean antisense RNA read density from the last (3′) 5 kb of all PTUs around a transcription stop site. The fifth block represents the mean antisense RNA read density from the last (3′) 5 kb of the 50 PTUs contiguous with convergent transcription termination regions lacking RNAP III transcribed genes. The antisense reads in the WT samples are in part derived from the authentic readthrough of the J-less convergent transcription termination region on chromosome 28 (see Figures 4 and 5) and in part from incidental antisense transcripts, probably due to mapping errors not yet corrected in the L. tarentolae genome. See also Table S5.
There is also an overall increase in antisense RNA in the L. tarentolae JBP2 KO (±BrdU) mutant that cannot be readily explained by readthrough or relaxed strand specificity at divergent transcription start regions. For example, chromosome 31 is transcribed (almost) exclusively from one strand but nevertheless displays an increase in antisense RNA (see Data S2). We return to this increase below.
Does Readthrough Lead to Accumulation of Duplex RNA?
The extensive readthrough at transcription stop sites when internal J levels fall could lead to formation of duplex RNA. As Leishmania does not have the machinery for processing of duplex RNA associated with RNA interference (Robinson and Beverley, 2003), any duplex RNA formed might accumulate. We have tested this by looking for ribonuclease (RNase)-resistant RNA, a time-honored method (Weissmann and Borst, 1963; Borst and Weissmann, 1965). None was found, although duplex viral RNA added as internal control was readily recovered (data not shown). Apparently, processing of nascent transcripts is fast enough to avoid duplex RNA formation, or Leishmania is able to rapidly degrade any duplex RNA formed by using enzymes other than those involved in RNA interference.
Effect of Readthrough on Sense RNA
Transcriptional readthrough has been shown to interfere with the synthesis of sense RNA in every prokaryotic and eukaryotic model system tested (Shearwin et al., 2005). To verify this for L. tarentolae, we analyzed the changes in the levels of sense RNA in the L. tarentolae JBP2 KO mutant grown in the presence or absence of BrdU relative to the WT RNA levels. No consistent change was found. However, we found changes in individual mRNA levels in the L. tarentolae JBP2 KO mutants spread over the entire genome (data not shown). Remarkably, mRNA levels (as measured by SL RNA-seq sense reads) in the mutant cells went both up and down, with 306 genes showing >2-fold decrease and 784 genes showing >2-fold increase SL RNA expression in the KO mutant relative to WT. The alterations in the KO+BrdU mutant were equally extensive.
We think that these widespread alterations are due not to the acute effects of J loss and readthrough but to secondary effects. After disruption of the second allele of the JBP2 gene, the mutant cells lose internal J slowly (Vainio et al., 2009), and after 15 passages, readthrough is still minimal (Figure S1). Only after at least 100 additional passages does the internal J level go down to the levels found here (Figure 2). This slow decrease of internal J provides the Leishmania mutant with ample time for adaptation, such as genome-wide increases and decreases in transcript levels. This transcriptional chaos makes it impossible to decide from our current data exactly how J loss kills Leishmania.
DISCUSSION
The function of base J has long been elusive (Borst and Sabatini, 2008), but the data presented here show that J is essential for proper termination of RNAP II transcription in Leishmania. J is present at all 38 convergent RNAP II transcription termination sites with only one exception. When one of the enzymes required for J biosynthesis, JBP2, is eliminated, the internal J peaks marking RNAP II termination sites decrease by an order of magnitude, and significant readthrough becomes detectable (Figure 2). This readthrough is exacerbated when the JBP2 KO cells are grown in BrdU. Under these conditions, the J peaks marking RNAP II termination sites disappear (nearly) completely (Figure 2).
The requirement of base J for RNAP II transcription termination is supported by the results obtained for the unique transcription termination site on chromosome 28 that is not marked by base J (Figure 5). At this site, we observe antisense RNA (interpreted as readthrough) in the WT cells, strongly indicating that proper termination of RNAP II transcription does not occur in the absence of base J unless RNAP III transcription meeting RNAP II stops the readthrough (Figures 3 and 4). We have scrutinized the one and only J-less readthrough region for proteins encoded in the antisense RNA, but we found none. We cannot rule out, however, that there might be useful noncoding RNAs derived from the antisense RNA, as readthrough at this convergent transcription termination region appears to be conserved in three other Leishmania species (L. donovani, L. amazonensis, and L. tarentolae) (P.J.M. and C.F., unpublished data). Readthrough at this site is not observed in L. braziliensis, however, which has an intact RNA interference system (Lye et al., 2010).
Many (26 out of 58 in L. tarentolae) divergent transcription start regions do not contain any detectable J (see Figure 1 for an example), showing that J is not required for transcription initiation in Leishmania. Nevertheless, transcription initiation is also affected when J is lost (Figure 2) because loss of J results in an increase of antisense SL-containing RNAs at transcription start sites (Figure 6). Although this increase in “false starts” is most pronounced at divergent transcription start regions containing J, it also occurs at divergent start regions that lack J, as well as at telomeric and “internal” transcription start sites. In addition, there appears to be a generalized low increase in antisense transcripts all over the genome (Figures 2 and 6). It is possible that there is always a low level of improper transcription initiation in WT cells but that these antisense transcripts are rapidly eliminated. Gross readthrough in the KO (±BrdU) mutants could overtax this system and could result in the appearance of increased antisense transcripts all over the genome. This hypothesis is in line with the observation that ~10% of transcripts detected in nuclear run-on experiments with L. major were antisense (Martínez-Calvillo et al., 2003) and with the presence of occasional antisense transcripts throughout the genome in WT cells (Belli et al., 2003). The overtaxing hypothesis could also explain the increased readthrough at the single convergent RNAP II termination site (28.2) not marked by J when J levels decrease (Figure 5). Finally, low levels of J, randomly located throughout the genome, could suppress rare but normally occurring antisense transcription. When this J is lost, a general increase of antisense RNA will occur. Although we have no evidence for this possibility, we cannot exclude it.
When cells lacking JBP2 are grown in BrdU, they lose additional J and die (Vainio et al., 2009). Our previous attempts to pinpoint the cause of death were unsuccessful (P.B., S.V., P.-A.G., and H.G.A.M.v.L., unpublished data). The massive readthrough demonstrated here may now provide an explanation for J-less death. Readthrough interferes with sense RNA synthesis in all systems studied thus far (reviewed in Shearwin et al., 2005), and it seems plausible that it will also do so in Leishmania.
How base J promotes RNAP II termination is not yet known. In other eukaryotes, polyadenylation of precursor mRNA initiates exonuclease activity by the exosome that will eventually overtake RNAP II and terminate transcription (reviewed by Richard and Manley, 2009). Leishmania, with its long polycistronic transcription units, cannot use polyadenylation-coupled termination, and it may therefore have recruited J as an alternative termination signal. We do not yet know how J is recognized by the termination machinery. A simple mechanism could involve the stalling of RNA polymerase at base J, resulting in discontinuation of RNA synthesis. Recent work on DNA synthesis has shown that some DNA polymerases may stall extensively at unusual bases in the template strand (Flusberg et al., 2010). Another obvious hypothesis is that JBP1, a thymidine hydroxylase that tightly binds to duplex J-DNA (Cross et al., 1999; Sabatini et al., 2002; Yu et al., 2007; Grover et al., 2007), could provide the termination signal. This hypothesis is made unattractive by our finding that there is not much JBP1 in the cell, only enough to cover ~1% of the existing base J (Toaldo et al., 2005). Another possibility is that the chromosome-internal regions with J cluster in the nucleus with telomeric repeats containing J. This could create a “silencing domain” through which RNAP II would not be able to proceed. Although telomeres are known to cluster in Leishmania (Dossin et al., 2008), there is no evidence that base J is involved in this process. We are testing whether chromosome-internal J colocalizes with telomeric J by using the methodology previously used to demonstrate clustering of telomeric repeats in Leishmania (Dossin et al., 2008).
Although the KO of JBP2 results in a substantial loss of J and concomitant readthrough, the loss is not complete. We therefore relied on a pharmacological trick—growth in BrdU—to further eliminate base J from transcription termination sites. BrdU interferes with J synthesis in Leishmania, T. brucei (van Leeuwen et al., 1998a), and T. cruzi (Ekanayake et al., 2011), but we don’t know how. JBP1 does not detectably bind to oligonucleotides containing BrdU, and the level of BrdU incorporation in the experiments presented here is relatively low—less than 1% replacement of T (P.B., S.V., P.-A.G., and H.G.A.M.v.L., unpublished data). We have recently shown that growth of WT L. tarentolae for 48 hr in a relatively high concentration of BrdU (10 μM) results in a uniform loss of J at all positions in the genome. Even broad, exotically shaped internal J peaks reduced by 80% do not alter shape (P.B. and H.G.A.M.v.L., unpublished data). A low T substitution by BrdU may therefore uniformly reduce J peaks. This suggests that the J-insertion machinery may require more than one unmodified T to act, even if J is already present in DNA. These results make it unlikely that the BrdU effect on J is due to the type of chromatin alterations found by BrdU incorporation in yeast, which is highly dependent on DNA sequence (Miki et al., 2010; Fujii et al., 2010).
Cliffe et al. (2010) have found J flanking RNAP II transcription units in T. brucei and T. cruzi and presented preliminary data that J is also present in this location in L. major. In a recent paper, they analyzed the transcriptional alterations in T. cruzi that follow the KO of JBP1 (which is viable in T. cruzi) or JBP2. They also find widespread alterations in transcription following J loss (Ekanayake et al., 2011), as we find in Leishmania, but no readthrough of RNAP II termination sites. Instead, there is a major increase in transcription initiation and a loosening of chromatin structure at divergent transcription start regions (Ekanayake et al., 2011). Whether there is also widespread synthesis of antisense RNA in T. cruzi was not tested. Leishmania and T. cruzi are distant relatives (Simpson et al., 2006; Stevens, 2008), comparable to frogs and apes. Hence, they may not use J for the same functions. However, the difference may seem larger than it is. We also see minor alterations at divergent start regions (Figures 2 and 6), even at starts regions not containing J. Suppression of transcription initiation and suppression of transcriptional readthrough may act through similar mechanisms in that they both could involve the creation of more repressive chromatin structures (Beisel and Paro, 2011). It is possible that transcription initiation is normally limited by association with telomeric clusters, as hypothesized here for transcription termination sites. If such nuclear repressive domains would be disrupted by loss of J, the precision of transcription initiation might decrease. RNAP II transcription initiation in Leishmania appears to be imprecise anyhow (Martínez-Calvillo et al., 2003, 2004), and DNA chromatin marks may be required to limit the position where the polymerase can start. Besides J, these marks could involve histone variants, which are known to mark the boundaries of polycistronic transcription units in trypanosomes (Siegel et al., 2009; Wright et al., 2010; Cliffe et al., 2010), and specific histone modifications (Thomas et al., 2009). The basic function of J may therefore be to help in the formation of repressive chromatin.
EXPERIMENTAL PROCEDURES
Detailed protocols are described in the Supplemental Information.
J-DNA Isolation and Quantification
Leishmania were grown in SDM-79 medium, and JBP2 KO cells (Vainio et al., 2009) were grown in 0.6 μM BrdU-containing medium for 5 days before harvesting. Genomic DNA was isolated and quantified by using immunoblots as described by van Leeuwen et al. (1997). For sequencing, the genomic DNA was dissolved in water, sheared (Covaris) to fragments of ~200 bp, and precipitated with anti-J antibodies (van Leeuwen et al., 1997). For precipitation with JBP1, biotin-tagged L. tarentolae JBP1 preattached to magnetic streptavidin-coated beads were used.
Cloning, Expression, and Purification of Biotin-Tagged L. tarentolae JBP1
The codon-optimized full-length L. tarentolae JBP1 gene (Heidebrecht et al., 2011) was used as a template to fuse a 16 residue N-terminal peptide tag required by the E. coli biotin ligase BirA for the enzymatic conjugation of a single biotin (Schatz, 1993). Tagged JBP1 was purified from E. coli by using streptavidin-coated magnetic beads (Dynabeads M-280, Invitrogen). The estimated concentration of JBP1 on the beads was 0.1–0.2 mg per ml.
High-Throughput Small RNA, SL RNA, dsRNA, and DNA Sequencing
Genomic DNA was sheared into fragments of 100–1,200 bp (Nebulizer, Illumina) and was made into a DNA sequencing (DNA-seq) library by using the Genomic DNA Sample Preparation Kit (Illumina, San Diego, USA). Chromatin immunoprecipitation sequencing (ChIP-seq) libraries were prepared with the Illumina sample preparation kit and modified in the use of Covaris DNA Shearing (Covaris, MA, USA). Small RNA was purified by using 15% polyacrylamide/urea/TBE gel electrophoresis and ligated to 5′ and 3′ small RNA adapters, reverse transcribed, and prepared for sequencing. SL RNA libraries were prepared by using an adapted protocol from Armour et al. (2009). Samples were sequenced by using an Illumina Genome Analyzer IIx.
Genome Annotation
Version 5.3 of the L. major Friedlin genome sequence and annotation was obtained from release 2.0 of TriTrypDB. The version 2 draft assembly of the L. tarentolae Parrot (LtaP) genome sequence was graciously provided by F. Raymond, B. Papadopoulou, M. Olivier, M.J. Tremblay, M. Ouellette, and J. Corbeil at CIHR in Québec, Canada (Raymond et al., 2012). Structural annotation was performed by reconciliation of three separate annotation efforts: the Rapid Annotation Transfer Tool (RATT) (Otto et al., 2011), the AutoMagi software (El-Sayed et al., 2005a), and Crossmatch (http://www.phrap.org/phredphrapconsed.html).
Mapping of High-Throughput Sequencing Reads and J Peaks
Small RNA reads were aligned by using the Burrows Wheeler Aligner (Li and Durbin, 2009) using the default parameters. ChIP-seq data were filtered to remove all reads with a mapping quality equal or less than 21. The small RNA data were filtered to remove the reads with a mapping quality value less than 37 or reads that aligned with mismatches. For all data sets, reads mapped to multiple sites in the genome were discarded. After mapping the J-IP sequence reads to the L. major and L. tarentolae genomes, the number of reads at each nucleotide position was used to define J peaks. A major peak was defined as contiguous sequence with a read depth of 20 or greater at each nucleotide and with a peak intensity of 100 or greater. Peaks due to collapsed repetitive sequence were usually ignored.
Calculation of Average Readthrough Profiles
Custom R scripts were used to generate average readthrough profiles. For this purpose, genome-wide positions of all read counts in 100 bp windows were converted to coordinates relative to the nearest transcription termination sites on the same strand, taking strand orientation of the reads and the corresponding PTU into account. Mean read counts across the aligned transcription termination sites were calculated with a running window covering 2% of the data.
Supplementary Material
ACKNOWLEDGMENTS
We thank F. Raymond, B. Papadopoulou, M. Olivier, M.J. Tremblay, M. Ouellette, and J. Corbeil at CIHR in Québec, Canada, for sharing the genomic sequence of L. tarentolae before publication. We thank U. Desselberger, University of Cambridge, UK, for providing purified rotavirus RNA preparations. We thank E. Christodoulou for help with JBP1 production. We are grateful to K. de Visser and F. van Leeuwen for critical reading of the manuscript and discussions. This work was supported by PHS grant R01 AI053667 from the National Institute of Allergy and Infectious Diseases to P.J.M.
Footnotes
ACCESSION NUMBERS
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE32976 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32976).
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, two data files, one figure, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.07.030.
REFERENCES
- Agabian N. Trans splicing of nuclear pre-mRNAs. Cell. 1990;61:1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- Armour CD, Castle JC, Chen R, Babak T, Loerch P, Jackson S, Shah JK, Dey J, Rohl CA, Johnson JM, Raymond CK. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat. Methods. 2009;6:647–649. doi: 10.1038/nmeth.1360. [DOI] [PubMed] [Google Scholar]
- Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat. Rev. Genet. 2011;12:123–135. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- Belli SI, Monnerat S, Schaff C, Masina S, Noll T, Myler PJ, Stuart K, Fasel N. Sense and antisense transcripts in the histone H1 (HIS-1) locus of Leishmania major. Int. J. Parasitol. 2003;33:965–975. doi: 10.1016/s0020-7519(03)00126-7. [DOI] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Boothroyd JC, Cross GAM. Transcripts coding for variant surface glycoproteins of Trypanosoma brucei have a short, identical exon at their 5′ end. Gene. 1982;20:281–289. doi: 10.1016/0378-1119(82)90046-4. [DOI] [PubMed] [Google Scholar]
- Borst P, Weissmann C. Replication of viral RNA, 8. Studies on the enzymatic mechanism of replication of MS2 RNA. Proc. Natl. Acad. Sci. USA. 1965;54:982–987. doi: 10.1073/pnas.54.3.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P, Sabatini R. Base J: discovery, biosynthesis, and possible functions. Annu. Rev. Microbiol. 2008;62:235–251. doi: 10.1146/annurev.micro.62.081307.162750. [DOI] [PubMed] [Google Scholar]
- Cliffe LJ, Kieft R, Southern T, Birkeland SR, Marshall M, Sweeney K, Sabatini R. JBP1 and JBP2 are two distinct thymidine hydroxylases involved in J biosynthesis in genomic DNA of African trypanosomes. Nucleic Acids Res. 2009;37:1452–1462. doi: 10.1093/nar/gkn1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe LJ, Siegel TN, Marshall M, Cross GA, Sabatini R. Two thymidine hydroxylases differentially regulate the formation of glucosylated DNA at regions flanking polymerase II polycistronic transcription units throughout the genome of Trypanosoma brucei. Nucleic Acids Res. 2010;38:3923–3935. doi: 10.1093/nar/gkq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross M, Kieft R, Sabatini R, Wilm M, de Kort M, van der Marel GA, van Boom JH, van Leeuwen F, Borst P. The modified base J is the target for a novel DNA-binding protein in kinetoplastid protozoans. EMBO J. 1999;18:6573–6581. doi: 10.1093/emboj/18.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross M, Kieft R, Sabatini R, Dirks-Mulder A, Chaves I, Borst P. J-binding protein increases the level and retention of the unusual base J in trypanosome DNA. Mol. Microbiol. 2002;46:37–47. doi: 10.1046/j.1365-2958.2002.03144.x. [DOI] [PubMed] [Google Scholar]
- De Lange T, Liu AY, Van der Ploeg LH, Borst P, Tromp MC, Van Boom JH. Tandem repetition of the 5′ mini-exon of variant surface glycoprotein genes: a multiple promoter for VSG gene transcription? Cell. 1983;34:891–900. doi: 10.1016/0092-8674(83)90546-9. [DOI] [PubMed] [Google Scholar]
- De Lange T, Berkvens TM, Veerman HJ, Frasch AC, Barry JD, Borst P. Comparison of the genes coding for the common 5′ terminal sequence of messenger RNAs in three trypanosome species. Nucleic Acids Res. 1984;12:4431–4443. doi: 10.1093/nar/12.11.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPaolo C, Kieft R, Cross M, Sabatini R. Regulation of trypanosome DNA glycosylation by a SWI2/SNF2-like protein. Mol. Cell. 2005;17:441–451. doi: 10.1016/j.molcel.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Dooijes D, Chaves I, Kieft R, Dirks-Mulder A, Martin W, Borst P. Base J originally found in kinetoplastida is also a minor constituent of nuclear DNA of Euglena gracilis. Nucleic Acids Res. 2000;28:3017–3021. doi: 10.1093/nar/28.16.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossin Fde.M., Dufour A, Dusch E, Siqueira-Neto JL, Moraes CB, Yang GS, Cano MI, Genovesio A, Freitas-Junior LH. Automated nuclear analysis of Leishmania major telomeric clusters reveals changes in their organization during the parasite’s life cycle. PLoS ONE. 2008;3:e2313. doi: 10.1371/journal.pone.0002313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekanayake DK, Minning T, Weatherly B, Gunasekera K, Nilsson D, Tarleton R, Ochsenreiter T, Sabatini R. Epigenetic regulation of transcription and virulence in Trypanosoma cruzi by O-linked thymine glucosylation of DNA. Mol. Cell. Biol. 2011;31:1690–1700. doi: 10.1128/MCB.01277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005a;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, Aggarwal G, Caler E, Renauld H, Worthey EA, Hertz-Fowler C, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005b;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, Miki K, Takayama S, Ayusawa D. Identification of genes that affect sensitivity to 5-bromodeoxyuridine in the yeast Saccharomyces cerevisiae. Mol. Genet. Genomics. 2010;283:461–468. doi: 10.1007/s00438-010-0535-6. [DOI] [PubMed] [Google Scholar]
- Genest PA, ter Riet B, Dumas C, Papadopoulou B, van Luenen HGAM, Borst P. Formation of linear inverted repeat amplicons following targeting of an essential gene in Leishmania. Nucleic Acids Res. 2005;33:1699–1709. doi: 10.1093/nar/gki304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest PA, Ter Riet B, Cijsouw T, van Luenen HG, Borst P. Telomeric localization of the modified DNA base J in the genome of the protozoan parasite Leishmania. Nucleic Acids Res. 2007;35:2116–2124. doi: 10.1093/nar/gkm050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers-Ampt JH, Van Leeuwen F, de Beer AL, Vliegenthart JF, Dizdaroglu M, Kowalak JA, Crain PF, Borst P. β-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993;75:1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- Grover RK, Pond SJ, Cui Q, Subramaniam P, Case DA, Millar DP, Wentworth P., Jr. O-glycoside orientation is an essential aspect of base J recognition by the kinetoplastid DNA-binding protein JBP1. Angew. Chem. Int. Ed. Engl. 2007;46:2839–2843. doi: 10.1002/anie.200604635. [DOI] [PubMed] [Google Scholar]
- Heidebrecht T, Christodoulou E, Chalmers MJ, Jan S, Ter Riet B, Grover RK, Joosten RP, Littler D, van Luenen H, Griffin PR, et al. The structural basis for recognition of base J containing DNA by a novel DNA binding domain in JBP1. Nucleic Acids Res. 2011;39:5715–5728. doi: 10.1093/nar/gkr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AP, Sanders M, Berry A, McQuillan J, Aslett MA, Quail MA, Chukualim B, Capewell P, MacLeod A, Melville SE, et al. The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human african trypanosomiasis. PLoS Negl. Trop. Dis. 2010;4:e658. doi: 10.1371/journal.pntd.0000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooter JM, Borst P. α-amanitin-insensitive transcription of variant surface glycoprotein genes provides further evidence for discontinuous transcription in trypanosomes. Nucleic Acids Res. 1984;12:9457–9472. doi: 10.1093/nar/12.24.9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye LF, Owens K, Shi H, Murta SM, Vieira AC, Turco SJ, Tschudi C, Ullu E, Beverley SM. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 2010;6:e1001161. doi: 10.1371/journal.ppat.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Calvillo S, Yan S, Nguyen D, Fox M, Stuart K, Myler PJ. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell. 2003;11:1291–1299. doi: 10.1016/s1097-2765(03)00143-6. [DOI] [PubMed] [Google Scholar]
- Martínez-Calvillo S, Nguyen D, Stuart K, Myler PJ. Transcription initiation and termination on Leishmania major chromosome 3. Eukaryot. Cell. 2004;3:506–517. doi: 10.1128/EC.3.2.506-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki K, Shimizu M, Fujii M, Takayama S, Hossain MN, Ayusawa D. 5-bromodeoxyuridine induces transcription of repressed genes with disruption of nucleosome positioning. FEBS J. 2010;277:4539–4548. doi: 10.1111/j.1742-4658.2010.07868.x. [DOI] [PubMed] [Google Scholar]
- Monnerat S, Martinez-Calvillo S, Worthey E, Myler PJ, Stuart KD, Fasel N. Genomic organization and gene expression in a chromosomal region of Leishmania major. Mol. Biochem. Parasitol. 2004;134:233–243. doi: 10.1016/j.molbiopara.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Myler PJ, Audleman L, deVos T, Hixson G, Kiser P, Lemley C, Magness C, Rickel E, Sisk E, Sunkin S, et al. Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc. Natl. Acad. Sci. USA. 1999;96:2902–2906. doi: 10.1073/pnas.96.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myler PJ, Sisk E, McDonagh PD, Martinez-Calvillo S, Schnaufer A, Sunkin SM, Yan S, Madhubala R, Ivens A, Stuart K. Genomic organization and gene function in Leishmania. Biochem. Soc. Trans. 2000;28:527–531. doi: 10.1042/bst0280527. [DOI] [PubMed] [Google Scholar]
- Nelson RG, Parsons M, Barr PJ, Stuart K, Selkirk M, Agabian N. Sequences homologous to the variant antigen mRNA spliced leader are located in tandem repeats and variable orphons in trypanosoma brucei. Cell. 1983;34:901–909. doi: 10.1016/0092-8674(83)90547-0. [DOI] [PubMed] [Google Scholar]
- Otto T, Dillon G, Degrave W, Berriman M. RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res. 2011;39:e57. doi: 10.1093/nar/gkq1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Mejía NE, Florencio-Martínez LE, Figueroa-Angulo EE, Manning-Cela RG, Hernández-Rivas R, Myler PJ, Martínez-Calvillo S. Gene organization and sequence analyses of transfer RNA genes in Trypanosomatid parasites. BMC Genomics. 2009;10:232. doi: 10.1186/1471-2164-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond F, Boisvert S, Roy G, Ritt J-F, Legare D, Isnard A, Stanke M, Olivier M, Tremblay MJ, Papadopoulou B, et al. Genome sequencing of the lizard parasite Leishmania tarentolae reveals loss of genes associated to the intracellular stage of human pathogenic species. Nucleic Acids Res. 2012;40:1131–1147. doi: 10.1093/nar/gkr834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson KA, Beverley SM. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol. Biochem. Parasitol. 2003;128:217–228. doi: 10.1016/s0166-6851(03)00079-3. [DOI] [PubMed] [Google Scholar]
- Sabatini R, Meeuwenoord N, van Boom JH, Borst P. Recognition of base J in duplex DNA by J-binding protein. J. Biol. Chem. 2002;277:958–966. doi: 10.1074/jbc.M109000200. [DOI] [PubMed] [Google Scholar]
- Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (N.Y.) 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB. Transcriptional interference—a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, Fenyo D, Wang X, Dewell S, Cross GA. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AG, Stevens JR, Lukes J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;22:168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Stevens JR. Kinetoplastid phylogenetics, with special reference to the evolution of parasitic trypanosomes. Parasite. 2008;15:226–232. doi: 10.1051/parasite/2008153226. [DOI] [PubMed] [Google Scholar]
- Sutton RE, Boothroyd JC. Evidence for trans splicing in trypanosomes. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, Green A, Sturm NR, Campbell DA, Myler PJ. Histone acetylations mark origins of polycistronic transcription in Leishmania major. BMC Genomics. 2009;10:152. doi: 10.1186/1471-2164-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toaldo CB, Kieft R, Dirks-Mulder A, Sabatini R, van Luenen HG, Borst P. A minor fraction of base J in kinetoplastid nuclear DNA is bound by the J-binding protein 1. Mol. Biochem. Parasitol. 2005;143:111–115. doi: 10.1016/j.molbiopara.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E, Matthews KR, Tschudi C. Temporal order of RNA-processing reactions in trypanosomes: rapid trans splicing precedes polyadenylation of newly synthesized tubulin transcripts. Mol. Cell. Biol. 1993;13:720–725. doi: 10.1128/mcb.13.1.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainio S, Genest PA, ter Riet B, van Luenen HGAM, Borst P. Evidence that J-binding protein 2 is a thymidine hydroxylase catalyzing the first step in the biosynthesis of DNA base. J. Mol. Biochem. Parasitol. 2009;164:157–161. doi: 10.1016/j.molbiopara.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Van der Ploeg LHT, Liu AYC, Michels PAM, De Lange T, Borst P, Majumder HK, Weber H, Veeneman GH, Van Boom J. RNA splicing is required to make the messenger RNA for a variant surface antigen in trypanosomes. Nucleic Acids Res. 1982;10:3591–3604. doi: 10.1093/nar/10.12.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Wijsman ER, Kuyl-Yeheskiely E, van der Marel GA, van Boom JH, Borst P. The telomeric GGGTTA repeats of Trypanosoma brucei contain the hypermodified base J in both strands. Nucleic Acids Res. 1996;24:2476–2482. doi: 10.1093/nar/24.13.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Wijsman ER, Kieft R, van der Marel GA, van Boom JH, Borst P. Localization of the modified base J in telomeric VSG gene expression sites of Trypanosoma brucei. Genes Dev. 1997;11:3232–3241. doi: 10.1101/gad.11.23.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Kieft R, Cross M, Borst P. Biosynthesis and function of the modified DNA base β-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol. Cell. Biol. 1998a;18:5643–5651. doi: 10.1128/mcb.18.10.5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Taylor MC, Mondragon A, Moreau H, Gibson W, Kieft R, Borst P. β-D-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres. Proc. Natl. Acad. Sci. USA. 1998b;95 doi: 10.1073/pnas.95.5.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Kieft R, Cross M, Borst P. Tandemly repeated DNA is a target for the partial replacement of thymine by β-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol. Biochem. Parasitol. 2000;109:133–145. doi: 10.1016/s0166-6851(00)00247-4. [DOI] [PubMed] [Google Scholar]
- Weissmann C, Borst P. Double-stranded ribonucleic acid formation in vitro by MS 2 phage-induced RNA synthetase. Science. 1963;142:1188–1191. doi: 10.1126/science.142.3596.1188. [DOI] [PubMed] [Google Scholar]
- Worthey EA, Martinez-Calvillo S, Schnaufer A, Aggarwal G, Cawthra J, Fazelinia G, Fong C, Fu G, Hassebrock M, Hixson G, et al. Leishmania major chromosome 3 contains two long convergent polycistronic gene clusters separated by a tRNA gene. Nucleic Acids Res. 2003;31:4201–4210. doi: 10.1093/nar/gkg469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JR, Siegel TN, Cross GA. Histone H3 trimethylated at lysine 4 is enriched at probable transcription start sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 2010;172:141–144. doi: 10.1016/j.molbiopara.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Genest PA, ter Riet B, Sweeney K, DiPaolo C, Kieft R, Christodoulou E, Perrakis A, Simmons JM, Hausinger RP, et al. The protein that binds to DNA base J in trypanosomatids has features of a thymidine hydroxylase. Nucleic Acids Res. 2007;35:2107–2115. doi: 10.1093/nar/gkm049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.