Three-dimensional cryoEM reconstruction of native LDL particles to 16Å resolution at physiological body temperature
- PMID: 21573056
- PMCID: PMC3090388
- DOI: 10.1371/journal.pone.0018841
Three-dimensional cryoEM reconstruction of native LDL particles to 16Å resolution at physiological body temperature
Abstract
Background: Low-density lipoprotein (LDL) particles, the major carriers of cholesterol in the human circulation, have a key role in cholesterol physiology and in the development of atherosclerosis. The most prominent structural components in LDL are the core-forming cholesteryl esters (CE) and the particle-encircling single copy of a huge, non-exchangeable protein, the apolipoprotein B-100 (apoB-100). The shape of native LDL particles and the conformation of native apoB-100 on the particles remain incompletely characterized at the physiological human body temperature (37 °C).
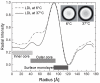
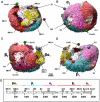
Methodology/principal findings: To study native LDL particles, we applied cryo-electron microscopy to calculate 3D reconstructions of LDL particles in their hydrated state. Images of the particles vitrified at 6 °C and 37 °C resulted in reconstructions at ~16 Å resolution at both temperatures. 3D variance map analysis revealed rigid and flexible domains of lipids and apoB-100 at both temperatures. The reconstructions showed less variability at 6 °C than at 37 °C, which reflected increased order of the core CE molecules, rather than decreased mobility of the apoB-100. Compact molecular packing of the core and order in a lipid-binding domain of apoB-100 were observed at 6 °C, but not at 37 °C. At 37 °C we were able to highlight features in the LDL particles that are not clearly separable in 3D maps at 6 °C. Segmentation of apoB-100 density, fitting of lipovitellin X-ray structure, and antibody mapping, jointly revealed the approximate locations of the individual domains of apoB-100 on the surface of native LDL particles.
Conclusions/significance: Our study provides molecular background for further understanding of the link between structure and function of native LDL particles at physiological body temperature.
Conflict of interest statement
Figures




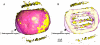

Similar articles
-
Immuno-electron cryo-microscopy imaging reveals a looped topology of apoB at the surface of human LDL.J Lipid Res. 2011 Jun;52(6):1111-1116. doi: 10.1194/jlr.M013946. Epub 2011 Apr 1. J Lipid Res. 2011. PMID: 21460103 Free PMC article.
-
Structure of triglyceride-rich human low-density lipoproteins according to cryoelectron microscopy.Biochemistry. 2003 Dec 23;42(50):14988-93. doi: 10.1021/bi0354738. Biochemistry. 2003. PMID: 14674775
-
Structure of apolipoprotein B-100 in low density lipoproteins.J Lipid Res. 2001 Sep;42(9):1346-67. J Lipid Res. 2001. PMID: 11518754 Review.
-
Human LDL core cholesterol ester packing: three-dimensional image reconstruction and SAXS simulation studies.J Lipid Res. 2011 Feb;52(2):256-62. doi: 10.1194/jlr.M011569. Epub 2010 Nov 3. J Lipid Res. 2011. PMID: 21047995 Free PMC article.
-
Apolipoprotein B and low-density lipoprotein structure: implications for biosynthesis of triglyceride-rich lipoproteins.Adv Protein Chem. 1994;45:205-48. doi: 10.1016/s0065-3233(08)60641-5. Adv Protein Chem. 1994. PMID: 8154370 Review.
Cited by
-
NMR spectroscopy of macrophages loaded with native, oxidized or enzymatically degraded lipoproteins.PLoS One. 2013;8(2):e56360. doi: 10.1371/journal.pone.0056360. Epub 2013 Feb 15. PLoS One. 2013. PMID: 23457556 Free PMC article.
-
Identification of a novel lipid binding motif in apolipoprotein B by the analysis of hydrophobic cluster domains.Biochim Biophys Acta Biomembr. 2017 Feb;1859(2):135-145. doi: 10.1016/j.bbamem.2016.10.019. Epub 2016 Nov 1. Biochim Biophys Acta Biomembr. 2017. PMID: 27814978 Free PMC article.
-
Plasminogen Deficiency Significantly Reduces Vascular Wall Disease in a Murine Model of Type IIa Hypercholesterolemia.Biomedicines. 2021 Dec 4;9(12):1832. doi: 10.3390/biomedicines9121832. Biomedicines. 2021. PMID: 34944648 Free PMC article.
-
Low-density lipoproteins investigated under high hydrostatic pressure by elastic incoherent neutron scattering.Eur Phys J E Soft Matter. 2017 Jul;40(7):68. doi: 10.1140/epje/i2017-11558-8. Epub 2017 Jul 26. Eur Phys J E Soft Matter. 2017. PMID: 28733727 Free PMC article.
-
Three dimensional electron microscopy and in silico tools for macromolecular structure determination.EXCLI J. 2013 Apr 24;12:335-46. eCollection 2013. EXCLI J. 2013. PMID: 27092033 Free PMC article. Review.
References
-
- Brown MS, Kovanen PT, Goldstein JL. Regulation of plasma cholesterol by lipoprotein receptors. Science. 1981;212:628–35. - PubMed
-
- Hegele RA. Plasma lipoproteins: genetic influences and clinical implications. Nat Rev Genet. 2009;10:109–121. - PubMed
-
- Williams KJ, Tabas I. The response-to-retention hypothesis of atherogenesis reinforced. Curr Opin Lipidol. 1998;9:471–474. - PubMed
-
- Hevonoja T, Pentikäinen MO, Hyvonen MT, Kovanen PT, Ala-Korpela M. Structure of low density lipoprotein (LDL) particles: basis for understanding molecular changes in modified LDL. Biochim Biophys Acta. 2000;1488:189–210. - PubMed
-
- Pentikäinen MO, Öörni K, Ala-Korpela M, Kovanen PT. Modified LDL - trigger of atherosclerosis and inflammation in the arterial intima. J Intern Med. 2000;247:359–370. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

